A bow to the mighty Python's C extension!
A little story how powerful Python’s C exstension are!
Note: I’m also learning. If you find any wrong or misleading information, please write to me.
Note: All the code you may find at my github in Blog repository
Hello after this long break!
I have got lots of work to do, but finally (at least for me) the new post is here!
Let me start from the beggining. Some time ago I have watched the Gynvaels video about genetic algorithm and drawning the Mona Lisa using it[1].
I was so amazed about the effect, that I have thought Hey, in Python it will be much easier! and started to work. Rather than Mona Lisa I went with The Lady with an Ermine gray:

So I wrote very simple, yet working script which I don’t paste to save the space. The whole script you may find here.
The results I got were very promissing (for 100, 300, 700 and 1000 generations, so the absolute minumum):

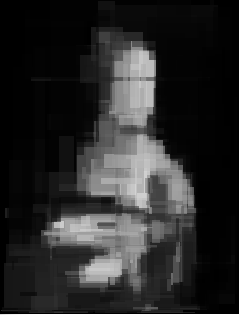
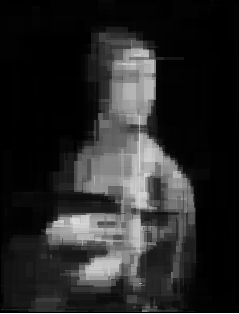

I only thought that drawning with triangles looks much more badass[2] so I tried to do it this way. If you don’t know how to render the triangle, don’t worry and read this.
And I must say I was 100% right! It looks awesome and that’s why this version of script is uploaded to my github. Just look at it!
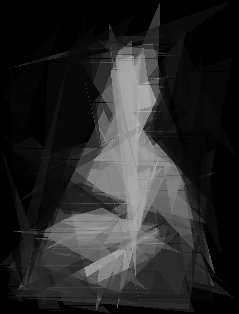


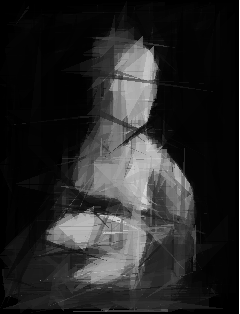
And I could end my story here, but there was one thing which bothered me the most. It was soooo slow! And I mean it. Possibly the slowest program I have ever written.
I made some test and generating 10 generations take around 137.5 seconds!
Can you imagine? For only 10 generations! To see some efects there must be at least 100 generations and 100 000 to get very good results. So I’m leaving the math to you.
I know what you’re thinking Python is slower than C and of course it is, but I did not know how much. One of the best programming languages must do better!
And beleive me, it does.
Let me introduce you to the Python's C extension!
As always the best way to learn something is to start with the documentation. As Python is very, very well documented, let’s dive into Extending and Embedding the Python Interpreter[3]. We can wrote that:
This document describes how to write modules in C or C++ to extend the Python interpreter with new modules. Those modules can not only define new functions but also new object types and their methods. The document also describes how to embed the Python interpreter in another application, for use as an extension language. Finally, it shows how to compile and link extension modules so that they can be loaded dynamically (at run time) into the interpreter, if the underlying operating system supports this feature.
I highly recommend you to read at least this introduction and after that we may start to write our extension. While testing the program I noticed the bottle-neck of the script is score function:
def score():
for i in range(POPULATION): #for every specimen in list of Specimens (fitness)
fitness[i].idx = i #assign current index
fitness[i].fitness_score = fitness[i].fitness(specimens[i]) #and count the fitness
fitness.sort(key = lambda x: x.fitness_score) #sort the fitness to get
#best results at the beginning
for i in range(BEST_ONES): #fill the bests with indexes of first BEST_ONES Specimens
bests[i] = fitness[i].idx
And I must say I’m not surprised, lots of things are happening there.
If this is the weak point I thought it is the best candidate to replace it with the C extension.
Please keep in mind this is not production code
I wrote the score module (score.c at my github), which implements three functions:
static PyObject* scoreFunc_score(PyObject* self, PyObject* args){
PyObject* specimensList;
PyObject* allSpec;
PyObject* art;
int W=0, H=0;
if (! PyArg_ParseTuple( args, "OiiOO", &specimensList, &W, &H, &allSpec, &art))
return NULL;
long specimen_lenght = PyList_Size(specimensList);
for(int i = 0; i < specimen_lenght; i++){
PyObject* tmp = PyList_GetItem(specimensList, i);
PyObject* m = Py_BuildValue("i", i);
PyObject_SetAttrString(tmp, "idx", m);
PyObject* ob2 = PyObject_GetAttrString(tmp, "idx");
idx = PyLong_AsLong(ob2);
PyObject* args_sM2 = Py_BuildValue("OiiiO", allSpec, W, H, idx, art);
res = scoreMe(args_sM2);
PyObject* set = Py_BuildValue("d", res);
PyObject_SetAttrString(tmp, "fitness_score", set);
PyObject* fitt = PyObject_GetAttrString(tmp, "fitness_score");
long fit = PyLong_AsLong(fitt);
}
return Py_None;
}
double scoreMe(PyObject* args){
PyObject* allSpec;
PyObject* art;
PyObject* final_score;
int W=0,H=0;
double sc = 0.0;
int idx;
if (! PyArg_ParseTuple( args, "OiiiO", &allSpec, &W, &H, &idx, &art))
return -1;
PyObject* tmp = PyList_GetItem(allSpec, idx);
for(int j = 0; j<H;j++){
for(int i = 0; i< W; i++){
long a = PyLong_AsLong(PyList_GetItem(tmp, j * W + i));
long b = PyLong_AsLong(PyList_GetItem(art, j * W + i));
sc += (a-b) * (a-b);
}
}
return PyLong_AsLong(Py_BuildValue("d", sc));
}
static PyObject* bestOnes(PyObject *self, PyObject* args){
PyObject* bestList;
PyObject* fitness;
int best_cnt;
if(!PyArg_ParseTuple(args, "OOi", &bestList, &fitness, &best_cnt))
return -1;
for(int i = 0; i<best_cnt; i++){
PyObject* tmp = PyList_GetItem(fitness, i);
int idx = PyLong_AsLong(PyObject_GetAttrString(tmp, "idx"));
PyObject* index = Py_BuildValue("i", idx);
PyList_SetItem(bestList, i, index);
}
return Py_None;
}
As you can see it is plain Cython!
I won’t be explainig this listing as everything can be found in Python/C API Reference Manual and the only thing you need to know is that those three functions do exactly the same what score function.
Next thing was to install our module and check if it works by importing it in Pythons terminal. If we manage to do that it is time to use it in our script. Type import score at the beginning of the file and let’s change the score function (I changed the function name to scoress to not to be mistaken while using the score function from score module).
def scoress():
score.score(fitness, W, H, specimens, picture)
fitness.sort(key = lambda x: x.fitness_score)
score.best_ones(bests, fitness, BEST_ONES)
As you can see I used my score and best_ones functions and did not bothered by implementing the sorting in Cython. In fact I would do that, but for now current result are enough.
So what are the results?!
I’m still impressed, so let me tell you. Generating 10 generations (as previous) using C extenstion took 8.8 seconds!. Yes! Only 8.8 seconds! It is almost 16 times less!
And implementing sorting would make it even faster!
Of course we have to remember that Python is very good and optimised language, but when you need some extra efficency, C extensions can provide it to you!
For today it is all! In next post I will cover the Python's Internals - part 2.
[1]. https://www.youtube.com/watch?v=7zI7M_5_jBE
[2]. https://rogerjohansson.blog/2008/12/07/genetic-programming-evolution-of-mona-lisa/
[3]. https://docs.python.org/3.6/extending/index.html#extending-index
